Introduction
A complex body of knowledge, such as science, is composed of various concepts and ideas that overlap and interconnect with one another. For example, in the discipline of physics, the concept of energy may be used to explain the motion of an object, whereas in chemistry, the energy concept may be used to explain the release of heat during a chemical reaction. In biology, the same concept may be used to illuminate the metabolic process within a cell. Therefore, the role of a teacher is to provide a thoughtful, well-designed curriculum, or a topical trajectory through many pieces of information, and help students to integrate different concepts toward a deep understanding of a topic.
A science textbook, with its sequence of chapters and sections, is an example of a carefully-designed trajectory through topics. While such reference sequences developed by subject experts are highly valuable, different topical trajectories can also be worthwhile and perhaps more effective for different teachers or students. Students may benefit from a different entry point to a topic based on their interest or background knowledge. For example, introductory physics may be taught in different sequences, based on whether a student is majoring in physics, chemistry, or biology. Different curricular sequences would highlight different connections between concepts and generate diverse insights about a topic.
We developed a method for producing topical sequences through a corpus of science textbooks, based on text analysis and path-minimizing heuristic algorithms. This method could be useful for students or teachers of introductory collegiate science courses, in which wide-ranging topics are introduced to a group of students whose interests and backgrounds are similarly wide-ranging. A teacher may use it to develop a lesson plan that explores connections between different topics and disciplines. A biology student who knows about the membrane potential of a cell from a biology course may benefit from reading about the concept of electrical potential from a physics textbook. Conversely, a physics student may reinforce the concept of electricity by learning about electrical phenomena from a biology textbook.
Finding informative terms
The texts from college-level introductory physics, chemistry, and biology textbooks from OpenStax (OpenStax College) were collected in plain text, ignoring special characters or mathematical symbols. Each section of the textbook is referred to as a “document” and is the smallest unit of data. The collection of these documents is referred to as a “corpus.” The punctuation marks and a set of common, uninformative words (such as “the”, “and”, or “a”) were removed, and the remaining words were transformed into their rudimentary forms or word stems, by the removal of suffixes like “-s” or “-ing.” For example, after stemming, “force”, “forces”, and “forced” are transformed into the same lexicon, “forc”. After a series of these preprocessing steps, each document was reduced into a collection of stemmed terms. These textural operations were performed with Python’s Natural Language ToolKit (NLTK) module (Bird, Loper and Klein, 2009).
Then, the term frequency-inverse document frequency (tf-idf) metric (Robertson and Spärck Jones, 1976) was calculated based on the frequency of each term within all documents in the corpus, according to the following formula:
tf-idf(t,d) = tf(t,d) x idf(t) = tf(t,d) x ( log( (1+N)/(1+df(t)) ) + 1 )
where tf(t,d) denotes the number of occurrences of each term (t) in each document (d), df(t) denotes the number of documents containing a particular term (t), and N is the total number of documents. The addition of 1 within the log function is to avoid division by zero or log of zero operation.
This metric quantifies the significance or the level of informativeness of a term within a corpus of documents. Low tf-idf value indicates that a term is not used often and/or it is a very generic term that appears throughout the entire corpus, so this term is not informative. High tf-idf value indicates that a term is used quite extensively in a certain document but not too often elsewhere within the corpus, so this term is likely an informative keyword for the document.
The following table shows a few examples of high tf-idf terms within a particular document from the textbook corpus. There is a reasonable correspondence between the topic of each document and these terms in their stemmed forms. A student or a teacher may use such a list to quickly identify important vocabularies of concepts within the documents.
| Section number and title (source: PHYS/CHEM/BIOL textbooks) |
Top 5 terms with large tf-idf values |
|---|---|
| PHYS 32.3
Therapeutic use of ionizing radiation |
cancer, radia, patient, tissu, ray |
| CHEM 6.2
Potential, kinetic, free, and activation energy |
energi, reaction, chemic, free, releas |
| BIOL 39.2
Gas exchange across respiratory surfaces |
volum, pressur, air, mm, oxygen |
The following table lists the top hundred informative terms, ranked by their average tf-idf values within physics, chemistry, and biology textbooks as well as the entire combined corpus. For example, in physics, “force” and “energy” are highly informative; in chemistry, “atom” and “electron”; and in biology, “cell” and “plant.” They represent keywords from three major branches of science.
| Corpus | Terms with highest tf-idf values |
|---|---|
| Physics | forc, energi, charg, wave, field, magnet, veloc, light, current, use, mass, motion, ray, object, electron, electr, two, direct, heat, one, point, temperatur, pressur, voltag, figur, momentum, acceler, time, particl, atom, speed, shown, water, imag, system, frequenc, show, resist, 10, radiat, vector, law, angl, wavelength, work, fluid, move, distanc, transfer, power, earth, sound, rotat, angular, chang, line, relat, calcul, produc, air, flow, posit, physic, circuit, kg, observ, exampl, equat, measur, given, nuclear, potenti, wire, surfac, zero, molecul, number, photon, differ, also, kinet, conserv, see, equal, valu, displac, 100, decay, first, find, part, right, length, small, exert, bodi, constant, reflect, per, weight |
| Chemistry | atom, bond, reaction, label, electron, aq, energi, acid, figur, two, water, solut, molecul, ion, use, subscript, carbon, right, compound, metal, contain, mass, rate, form, structur, one, 10, orbit, pressur, concentr, arrow, group, gas, temperatur, element, show, oxid, equilibrium, chemic, sphere, number, point, solid, shown, left, liquid, cell, exampl, chang, hydrogen, superscript, oxygen, line, process, equat, heat, particl, volum, red, substanc, negat, mol, pair, diagram, reactant, tube, charg, amount, first, three, sign, law, imag, valu, unit, work, properti, ionic, chapter, calcul, second, axi, column, blue, increas, product, spontan, formula, molecular, measur, singl, solubl, constant, follow, base, oh, credit, green, relat, modif |
| Biology | cell, plant, speci, figur, organ, protein, energi, dna, anim, gene, water, form, bodi, molecul, blood, system, show, use, popul, two, one, chromosom, call, membran, human, work, acid, hormon, produc, credit, carbon, also, differ, tissu, modif, structur, develop, group, reproduct, prokaryot, may, process, cycl, diseas, food, bind, mani, live, photo, tree, flower, life, bacteria, genet, includ, egg, function, part, environ, bone, fungi, receptor, exampl, oxygen, reaction, caus, virus, rna, signal, root, eukaryot, infect, activ, transcript, releas, type, light, result, chang, occur, contain, scientist, glucos, seed, individu, male, sequenc, enzym, atp, genom, like, fish, increas, respons, muscl, growth, electron, femal, regul, fertil |
| Physics + Chemistry + Biology | cell, energi, forc, atom, figur, use, plant, two, water, electron, one, molecul, reaction, organ, show, mass, system, speci, form, light, charg, bond, acid, protein, wave, veloc, temperatur, work, pressur, label, point, magnet, anim, field, bodi, dna, produc, gene, carbon, shown, heat, blood, structur, time, current, object, 10, chang, exampl, direct, particl, electr, call, differ, ion, ray, also, process, group, contain, right, motion, credit, human, solut, number, modif, imag, membran, mani, law, line, speed, relat, may, move, rate, air, equat, gas, increas, oxygen, calcul, arrow, part, compound, chemic, surfac, acceler, develop, left, earth, first, radiat, three, popul, food, result, subscript, frequenc |
A Venn diagram of these key terms provides a way to visualize the distinctive and overlapping concepts between three major branches of science. The terms like “reaction” and “cell” are found at the intersection between biology and chemistry, and the terms like “atom” and “temperature” are at the intersection between chemistry and physics. Overlapping terms between biology and physics are fewer and tend to have lower tf-idf values. At the intersection of all three disciplines, there are such nouns as “energy”, “electron”, and “water.” The overlapping term “change” is also interesting, because scientists in all disciplines describe many important natural phenomena in terms of changes in various quantities or properties: changes in temperature, molecular structure, environments, energy, etc. The word “work” was most often used as a citation terminology in the corpus (i.e., “work by so-and-so”). Other expository terms like “example”, “figure”, “show”, and “use” are understandably used quite often across all three textbooks.


A breakdown of tf-idf values within physics, chemistry, and biology textbooks is shown in Figure 2 for selected key terms. The terms “cell” and “plant” have particularly high tf-idf value within biology. The terms “energy” and “force” are significant within physics, and the terms “atom” and “reaction” are significant within chemistry. These results are quite reasonable given the general focus of each discipline and indicate that tf-idf is an acceptable metric of topical significance in the corpus of science textbooks.
By considering the correlation of tf-idf values across documents, it is possible to examine which terms are closely related to one another. This examination further reveals disciplinary differences. For example, the term “energy” is highly correlated with terms like “kinetic”, “conserved”, “work”, “electron”, and “potential” within physics, while it is correlated with terms like “heat”, “acid”, “reaction”, “electron”, and “ion” within chemistry, and “reaction”, “molecule”, “cell”, “gene”, and “metabolism” within biology. The term “water” goes with “pressure”, “temperature”, “density”, “fluid”, and “heat” in physics; “liquid”, “atom”, “solution”, “electron”, and “bond” in chemistry; and “plant”, “gene”, “DNA”, “ocean”, and “cell” in biology.
An interesting term is “vector” which in physics refers to a mathematical quantity with direction and magnitude and in biology means a vehicle for delivering genetic material. Hence, in the physics textbook, it is correlated with terms like “force” (a vector quantity), “direction”, and “magnitude”, and in the biology textbook, it is correlated with “disease”, “DNA”, “transmit”, and “carry”.
| Physics | Chemistry | Biology | |
|---|---|---|---|
| Energy | kinet, conserv, work, potenti, magnet | heat, acid, reaction, electron, ion | reaction, molecul, cell, gene, metabol |
| Atom | electron, nucleus, molecul, forc, veloc | bond, electron, reaction, structur, solut | carbon, bond, molecul, electron, hydrogen |
| Water | pressur, temperatur, fluid, densiti, heat | liquid, atom, solut, electron, bond | plant, gene, dna, ocean, cell |
| Electron | atom, particl, charg, energi, wavelength | atom, orbit, bond, reaction, pair | reaction, atom, molecul, energi, hydrogen |
| Vector | forc, direct, magnitud, energi, point | bond, pair, electron, structur, atom | diseas, human, dna, transmit, carri |
As illustrated in Table 1, each document in a corpus can be represented by a set of tf-idf values. Section 32.3 “Therapeutic use of ionizing radiation” can be described as having high tf-idf value for the term “radia” but low tf-idf for “oxygen”. Conversely, Section 39.2 “Gas exchange across respiratory surfaces” has low tf-idf for “radia” but high tf-idf for “oxygen.” Furthermore, the pattern of tf-idf values for a fixed set of vocabulary can reveal the topical relationship between documents. For example, another document on the topic of gas would have a similar pattern of tf-idf values as Section 39.2.



Given such quantitative representation of each document, the corpus of science textbooks can be analyzed with statistical techniques, such as Principal Component Analysis for reducing the dimensionality of the dataset into two dimensions for visualization (Jolliffe and Cadima, 2016; Pedregosa et al., 2011; Hinton and Salakhutdinov, 2006). When the entire dataset is rendered as a two-dimensional scatterplot, where each data point corresponds to an individual section and where the position of each point is determined by the principal components of tf-idf values, it is readily apparent that there are three distinct clusters, corresponding to physics, chemistry, and biology sections.


Navigating through the documents
Once each document is vectorized with tf-idf values, the documents can be considered as points in a high-dimensional space or a node in a graph. The similarity, or the length of an edge, between two documents can be calculated as a pairwise distance with a Euclidean metric, for example. A total distance of a path containing multiple nodes is the sum of pairwise distances. The situation is akin to the Traveling Salesman Problem (TSP), where the objective is to minimize the traveling distance while visiting multiple cities. However, since there are many nodes (~600) in our data, it is computationally prohibitive to search for a global optimum out of ~600 factorial possible paths. Furthermore, the goal here is to explore different paths through documents and to discover interesting curricular paths, rather than to find a single best path.
A simple heuristic of hopping from the current node to the next nearest node, as long as it has not been visited before, can produce a well-determined path. Because the nearby nodes, by construction, have the similar tf-idf values, they are the documents that contain similar terms and deal with related topics. Therefore, when one navigates through such a path, there will not likely be few conceptual leaps or gaps. However, this heuristic alone is not capable of exploring diverse paths. The following stochastic process, similar to a simulated annealing algorithm (Kirkpatrick, Gelatt Jr, and Vecchi, 1983), can be added: the next nearest node may be accepted with the probability p (or the nearest node is rejected with the probability 1-p). Then, this algorithm could discover a better global trajectory, even though it contains locally less optimal path segments, as shown in the following figure.

The following is an example of different paths explored by the algorithm, which start at the same document (on the topic of electric potential from the physics textbook), follow different trajectories and finish at the same document (on neural communication from the biology textbook).
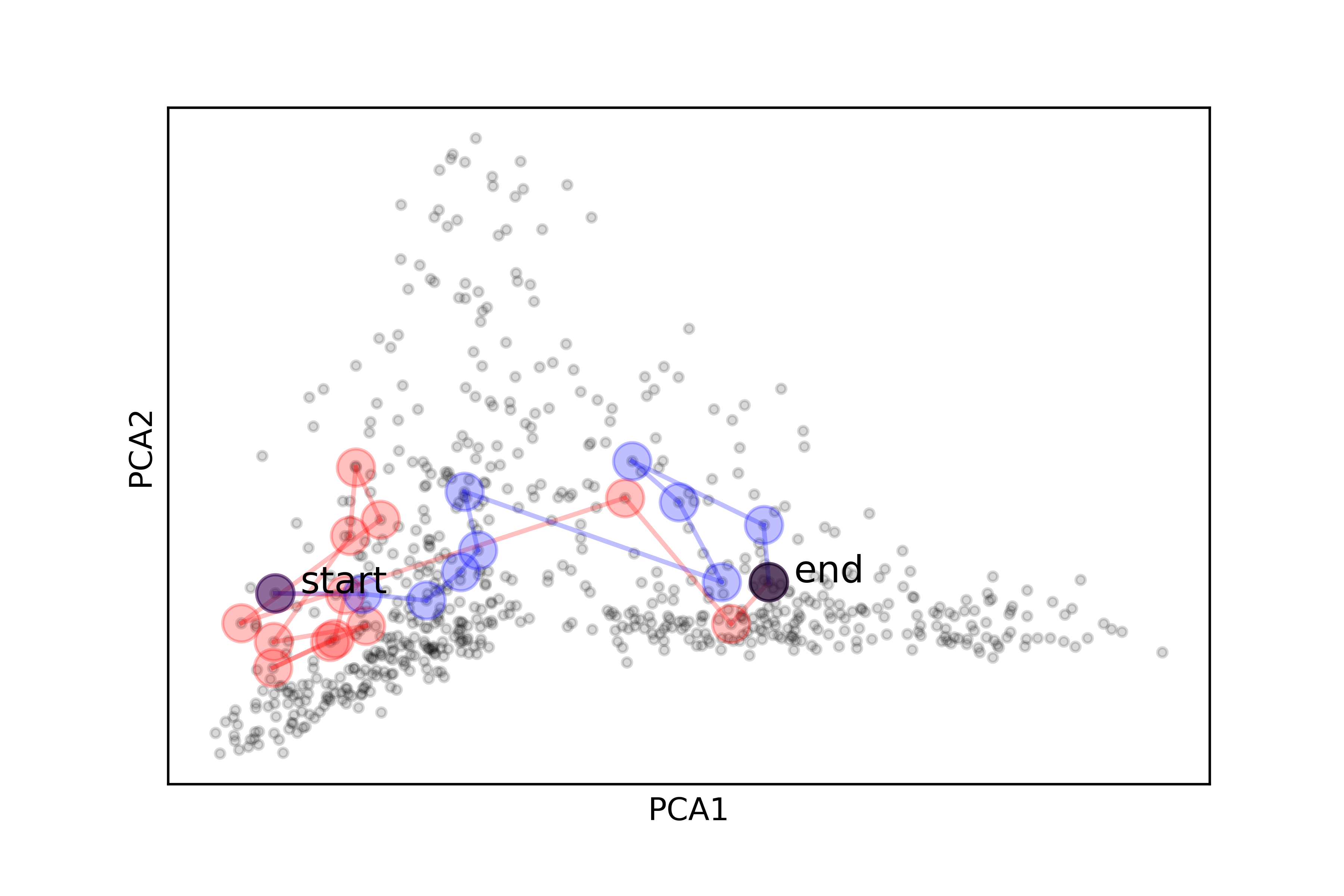

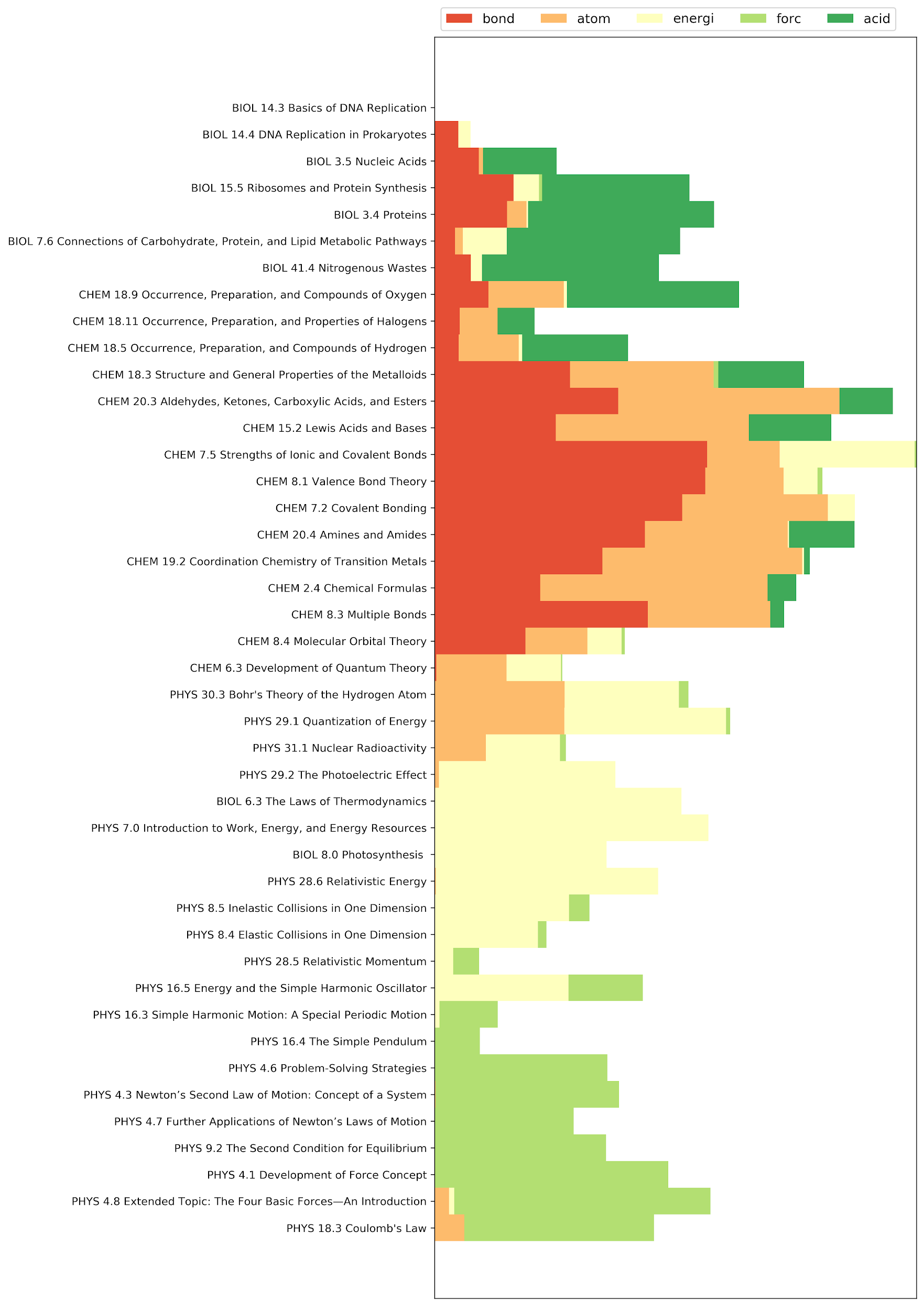
One may further analyze the paths by examining the terms with significant changes in the tf-idf values. The following figure displays the document titles of two different sample paths explored by the algorithm, along with the tf-idf values of selected terms. These terms have the largest average tf-idf values in the paths. It is noteworthy that, although these paths start and end at the same points, they involve different sets of terms and concepts. The first sample path connects the documents mostly within the physics corpus and relies heavily on the topic of electrical charge and potential. More biology documents show up in the second sample path, and some of the significant terms in those documents are energy and cell.



Discovering multidisciplinary topics
The pairwise distance between documents can be used to examine the neighborhood of a given document and the level of interdisciplinarity. A document that is close to other documents from different disciplines can be regarded as containing a highly interdisciplinary topic. For example, the neighborhood of the following documents contains other documents from biology, chemistry, and physics. More specifically, each of these documents had at least two documents from each of three textbooks among its thirteen closest neighbors. Other selection criteria of different neighborhood sizes produced similar results.
| Topic | Nearest Neighbors |
|---|---|
| PHYS: 32.3 Therapeutic Uses of Ionizing Radiation | PHYS: 32.2 Biological Effects of Ionizing Radiation CHEM: 21.6 Biological Effects of Radiation PHYS: 31.1 Nuclear Radioactivity BIOL: 16.7 Cancer and Gene Regulation PHYS: 32.1 Medical Imaging and Diagnostics CHEM: 21.5 Uses of Radioisotopes PHYS: 31.2 Radiation Detection and Detectors PHYS: 29.3 Photon Energies and the Electromagnetic Spectrum BIOL: 17.4 Applying Genomics BIOL: 9.3 Response to the Signal PHYS: 14.7 Radiation CHEM: 21.3 Radioactive Decay PHYS: 24.3 The Electromagnetic Spectrum |
| BIOL: 6.2 Potential, Kinetic, Free, and Activation Energy | CHEM: 5.0 Thermochemistry PHYS: 7.6 Conservation of Energy BIOL: 6.3 The Laws of Thermodynamics BIOL: 6.4 ATP: Adenosine Triphosphate BIOL: 6.0 Metabolism BIOL: 6.1 Energy and Metabolism PHYS: 7.0 Introduction to Work, Energy, and Energy Resources PHYS: 32.5 Fusion CHEM: 12.5 Collision Theory CHEM: 5.3 Enthalpy CHEM: 16.4 Free Energy BIOL: 7.1 Energy in Living Systems BIOL: 8.1 Overview of Photosynthesis |
| BIOL: 39.2 Gas Exchange across Respiratory Surfaces | BIOL: 39.3 Breathing CHEM: 9.3 Stoichiometry of Gaseous Substances, Mixtures, and Reactions CHEM: 9.2 Relating Pressure, Volume, Amount, and Temperature: The Ideal Gas Law PHYS: 11.9 Pressure in the Body PHYS: 11.6 Gauge Pressure, Absolute Pressure, and Pressure Measurement BIOL: 39.4 Transport of Gases in Human Bodily Fluids PHYS: 13.3 The Ideal Gas Law CHEM: 9.1 Gas Pressure BIOL: 39.1 Systems of Gas Exchange PHYS: 13.6 Humidity, Evaporation, and Boiling PHYS: 13.5 Phase Changes BIOL: 40.4 Blood Flow and Blood Pressure Regulation PHYS: 11.4 Variation of Pressure with Depth in a Fluid |
| BIOL: 44.5 Climate and the Effects of Global Climate Change | BIOL: 46.3 Biogeochemical Cycles PHYS: 14.3 Phase Change and Latent Heat PHYS: 13.2 Thermal Expansion of Solids and Liquids BIOL: 44.2 Biogeography BIOL: 27.4 The Evolutionary History of the Animal Kingdom PHYS: 13.1 Temperature PHYS: 14.7 Radiation BIOL: 22.3 Prokaryotic Metabolism CHEM: 10.4 Phase Diagrams PHYS: 13.5 Phase Changes CHEM: 10.3 Phase Transitions CHEM: 18.6 Occurrence, Preparation, and Properties of Carbonates BIOL: 8.3 Using Light Energy to Make Organic Molecules |
These interdisciplinary documents deal with the topics of radioactivity, energy, breathing, and climate change. The list of neighbors hints at possible multidisciplinary science curriculum. For example, a curriculum on radiation could involve a discussion about the physical attributes of radiation (e.g., photon energy and nuclear physics) as well as about its impact on biology and medicine (e.g., radiation therapy or radioisotope imaging) and chemistry (e.g., radioisotopes). A lesson on breathing could cover the topics of respiratory organ, property of gas, and physical characterization of pressure and temperature.
Reproducible codes and step-by-step guides
The Python codes and the data of the tf-idf values are available at GitHub.
There are three Jupyter Notebooks and one master data file (OS_all_M_T_title.p). The first notebook (step1_txt2tfidf_data.ipynb) reads in a set of raw text files (individual sections of the science textbooks), calculates the tf-idf values and performs statistical analysis such as Principal Component Analysis. It also saves the calculated results with a filename OS_all_M_T_title.p. The second notebook (step2_tfidf2seq.ipynb) uses the saved tf-idf values from the previous analysis, implements the path-minimizing heuristic, and produces the sequence of sections.
The last notebook (step3_user_interface) can be run as a standalone code, as long as the data file (OS_all_M_T_title.p) is present. It allows a user to build different curricular sequences based on OpenStax textbooks, by entering various starting and ending points from drop-down menus and experimenting with other parameters in the algorithm. This notebook also creates an interactive 3D scatter plot as a standalone html file (scatter_3D.html).


Discussion and further considerations
Natural science is broadly and imprecisely divided into three categories: physics, chemistry, and biology. Although such categorization provides a useful framework for studying the vast world of science, it is merely a rough characterization that could unfortunately obstruct the broad applicability and interrelatedness of fundamental concepts across disciplines. It is therefore important in science education to emphasize the inter- and cross-disciplinary nature of science.
In this study, we used statistical and computational techniques to analyze college-level, introductory science textbooks. Our results show that there certainly are disciplinary distinctions between physics, chemistry, and biology textbooks, but at the same time, there are multidisciplinary topics and concepts that are shared across disciplines. Radioactivity, energy, breathing, and climate change were found to be particularly conducive to cross-disciplinary lessons. We have also introduced a heuristic algorithm for generating a sequence of reading, which may be used to develop interesting, nonlinear lesson plans that are different from the linear sequence of the original textbook. It may further be used to augment the traditional curriculum and expand the curricular scope to include other disciplines. The above visualizations of science textbooks could provide students with a multidisciplinary perspective, too.
The codes for reproducing our results are made available on GitHub, and these codes may be adapted to analyze texts from other disciplines, such as social sciences and humanities. Similar approaches may be further applied to legal or healthcare documents (Hobson, 2019), while it should also be recognized that there are biases in data and limitations in algorithms (O’Neal, 2016). For example, if a certain topic is either under- or over-emphasized in the corpus of texts, the results will inherit similar biases. Our simple heuristic algorithm has the advantage of being simple and interpretable, but, because of its simplicity, its outputs are not always usable. Many of the paths that are suggested by the algorithm may not be viable as a curricular sequence, so they should be inspected by an experienced instructor before being deployed to a student body.
Bibliography
Bird, Steven, Ewan Klein, and Edward Loper. 2009. Natural Language Processing with Python. Sebastopol, California: O’Reilly.
Hinton, Geoffrey E., and Ruslan R. Salakhutdinov. 2006. “Reducing the Dimensionality of Data with Neural Networks.” Science 313, no. 5786 (July 28): 504–7. https://doi.org/10.1126/science.1127647.
Hodson, Richard. 2019. “Digital Health.” Nature S97, 573, no. 7775 (September 25). https://doi.org/10.1038/d41586-019-02869-x.
Jolliffe, Ian T., and Jorge Cadima. 2016. “Principal Component Analysis: a Review and Recent Developments.” Philosophical Transactions of the Royal Society A: Mathematical, Physical and Engineering Sciences 374, no. 2065 (April 13). https://doi.org/10.1098/rsta.2015.0202.
Kirkpatrick, Scott, Charles Daniel Gelatt, Jr., and Mario P. Vecchi. 1983. “Optimization by Simulated Annealing.” Science 220, no. 4598 (May 13): 671–80. https://doi.org/10.1126/science.220.4598.671.
O’Neil, Cathy. 2016. Weapons of Math Destruction: How Big Data Increases Inequality and Threatens Democracy. New York: Crown Publishing.
OpenStax College. 2018. Biology 2e. Accessed June, 2019. https://openstax.org/details/books/biology-2e.
OpenStax College. 2018. Chemistry 2e. Accessed June, 2019. https://openstax.org/details/books/chemistry-2e.
OpenStax College. 2018. College Physics. Accessed June, 2019. https://openstax.org/details/books/college-physics.
Pedregosa, Fabrice, et al. 2011. “Scikit-learn: Machine Learning in Python.” Journal of Machine Learning Research 12: 2825–2830.
Robertson, Stephen. E., and Karen Sparck Jones. 1976. “Relevance Weighting of Search Terms.” Journal of the American Society for Information Science 27, no. 3: 129–46. https://doi.org/10.1002/asi.4630270302.
Acknowledgments
This project was supported by Drew University’s Digital Humanities research grant from Andrew W. Mellon Foundation.





'Constructing an Introductory Science Curriculum with Text Analysis and a Path-Minimizing Heuristic Algorithm' has 3 comments
September 6, 2022 @ 9:38 am josefkook846
The Microsoft Authenticator app allows you to login into your accounts when two-step verification is enabled. Passwords can be lost, stolen, or compromised; thus, two-step verification will allow you to access your accounts more securely. Aka.ms/authapp
e
July 20, 2022 @ 12:11 pm donalderic858
BJ’s Wholesale Club Holdings, Inc., commonly referred to as BJ’s, is an American membership-only warehouse club chain founded on February 6, 1984; 38 years ago. BJ’s Headquarters is in Westborough, MA, the United States which has 229 locations all over the Eastern United States, Michigan, and Ohio.
Bjs Allergen Menu 2022
June 21, 2022 @ 5:33 am Heed
Nice post. I am paramedic student. https://heedhealtheducation-australia.blogspot.com/2022/04/how-to-become-paramedic-heed-health.html